AMDAT
find_between
find_between creates a trajectory of particles (from the first target) that reside between two other particles (from the second target) subject to constraints on the total distance and angle formed between the two vectors.
find_between <name of trajectory list to create> <dist_cutoff> <costheta_cutoff> <is_only_diff>
<target1>
<target2>
2D illustration of find_between command with two particles separated by a distance of 3</a>
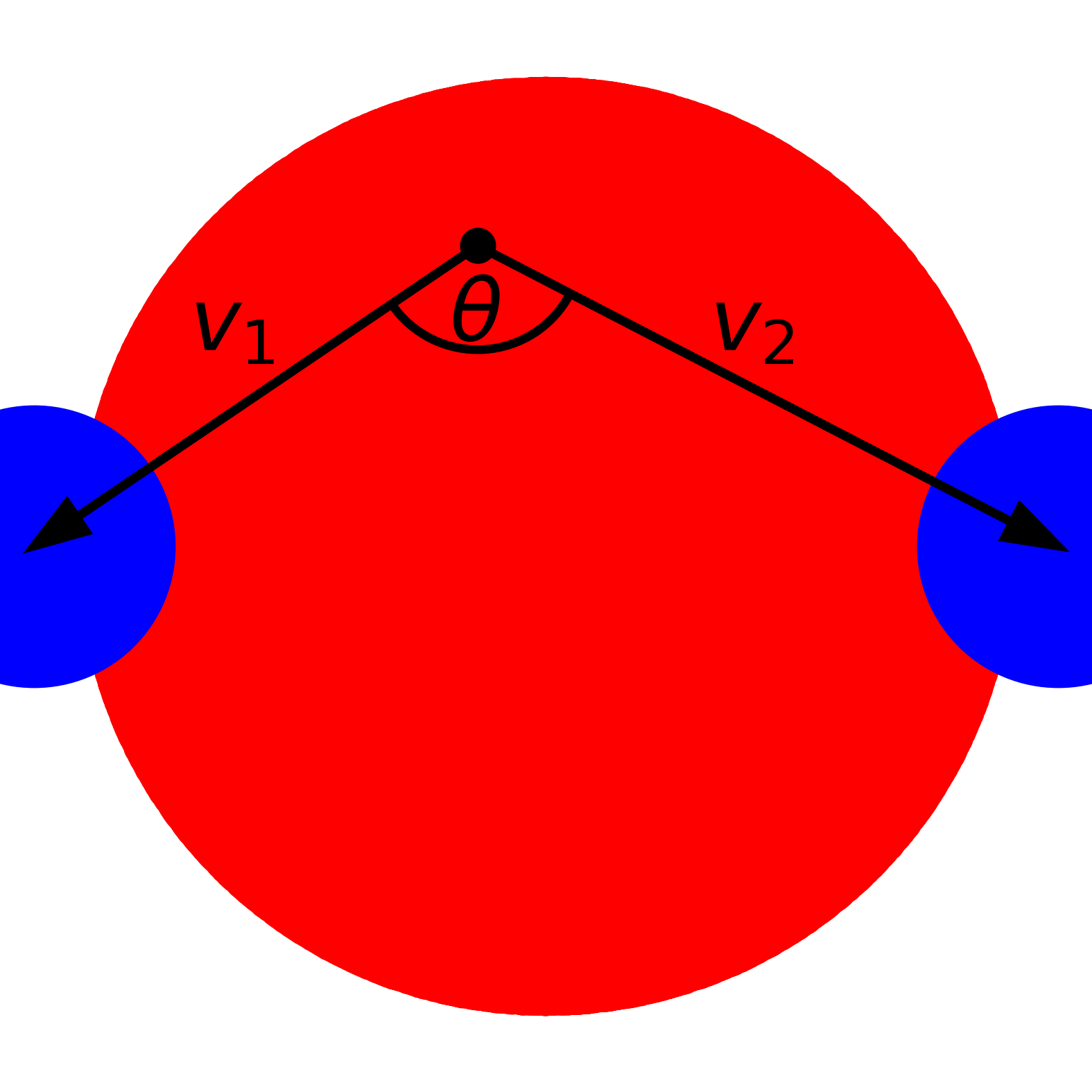
dist_cutoff = 4.0;costheta_cutoff = 0.0;Vectors and angle for a sample point |
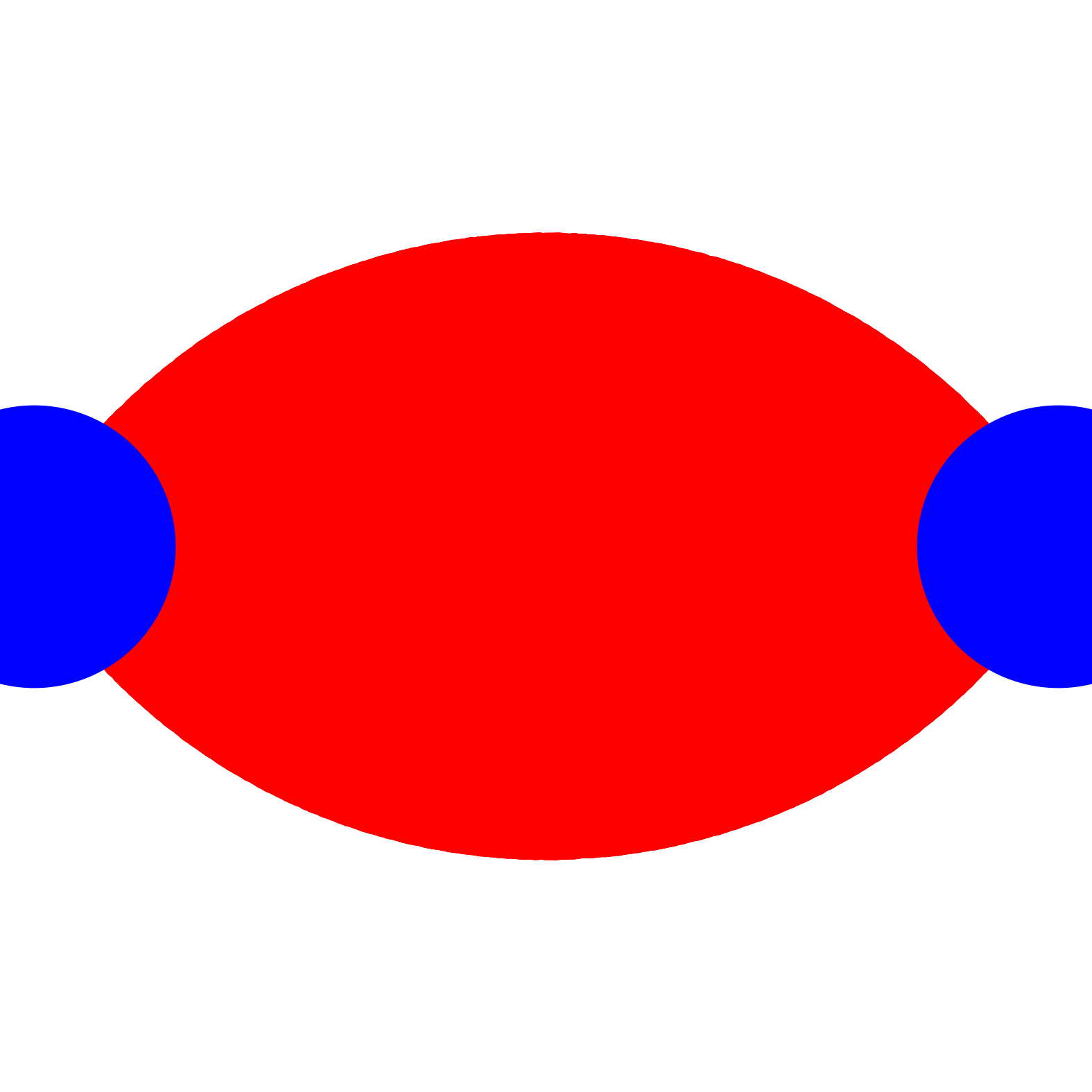
dist_cutoff = 4.0;costheta_cutoff = -0.5
|
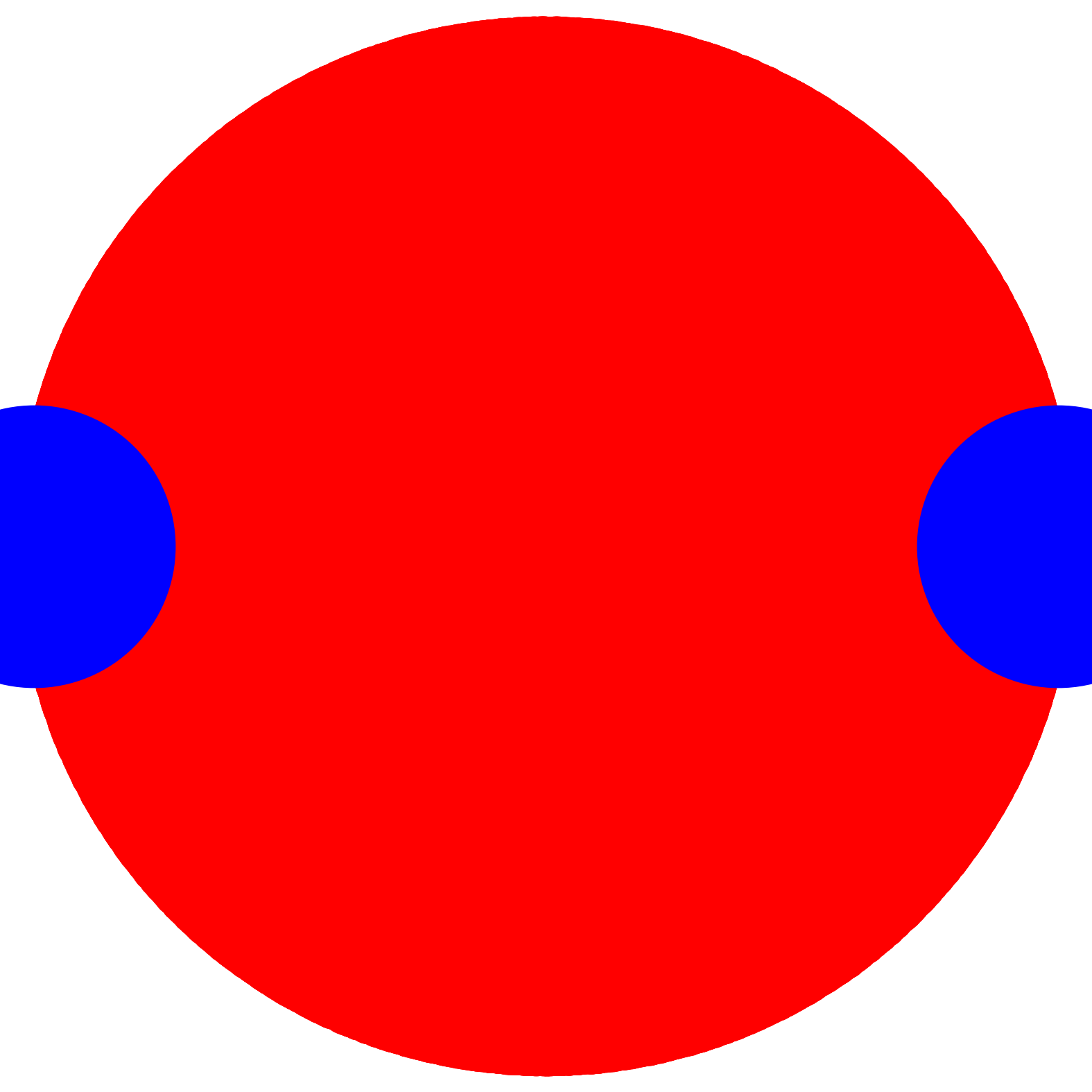
dist_cutoff = 5.0;costheta_cutoff = 0.0
|
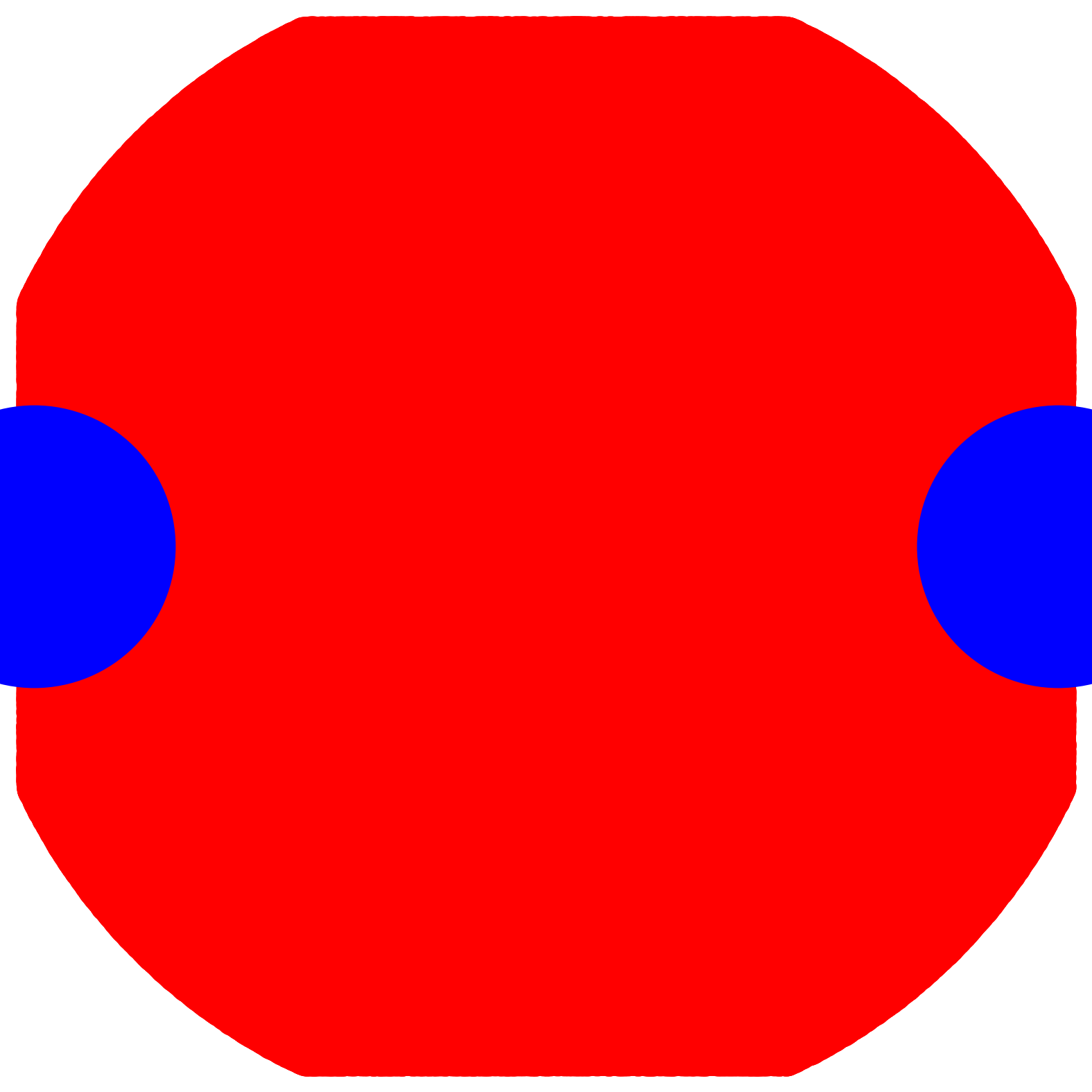
dist_cutoff = 5.0;costheta_cutoff = 0.5
|
Developed for Kawak, Bhapkar, Simmons. Macromolecules 57 (2024). doi: 10.1021/acs.macromol.4c00489
As can be seen by the illustration above, AMDAT will compute two vectors with an atom from the first target (<target1>) as the center with two atoms from the second target (<target2>).
An atom from the first target will be added to the resulting trajectory list if the two vector magnitudes ($d_1$ and $d_2$) and the angle formed between them obey ($\theta$) the following criteria:
\(\frac{d_1+d_2}{2} < d_c\) \(\cos \theta < \cos \theta_c\)
where $d_c$ is <dist_cutoff> and $\cos \theta_c$ is <costheta_cutoff>. The final option in this command is <is_only_diff> and it ensures that the two atoms from the second target do not belong to the same molecule. This last option helps distinguish inter-molecular phenomena from intra-molecular phenomena.